Exosomal Proteomics and Lipidomics Services
Creative Bioarray provides exosomal proteomics and lipidomics services based on mass spectrometry. Lipids and proteins are important components of biological exosomes, especially proteins. High-throughput analysis from omics helps researchers to identify and analyze the protein and lipid components of exosomes. The acquired knowledge can help understand the function of exosomes and discover new potential targets.
High-throughput Analysis of Exosomes
The molecular composition of extracellular vesicles (EV) such as exosomes is simple, but each type of molecule has thousands of variations. The development of omics technology is necessary for the high-throughput analysis of biomolecules. Single-objective analysis has a limited field of vision and is obviously inferior to global analysis in understanding biological events. In order to decipher the molecular mechanisms of vesicle cargo sorting and biogenesis, high-throughput analysis of EV protein and lipid components in various cell types and body fluids has become a popular trend. The common vesicle subsets of proteins, mRNA, miRNA, and lipids provide us with some important clues about the molecular mechanisms of vesicle cargo sorting and biogenesis. The cell-type-specific vesicle cargo helps us understand the pathophysiological functions outside the cell. A systematic analysis of the composition of vesicles shows that extracellular vesicles contain specific subsets of protein, genetic material (mRNA and miRNA), and lipids, rather than random cellular components. Transcriptomics, proteomics, and lipidomics based on high-throughput mass spectrometry facilitate the analysis of RNA-protein, protein-protein interactions in EVs, and strengthen the interaction analysis of signal pathways. For example, the protein-protein interaction network analysis of extracellular vesicles provides us with important clues about the molecular mechanism of vesicle cargo sorting and biogenesis.
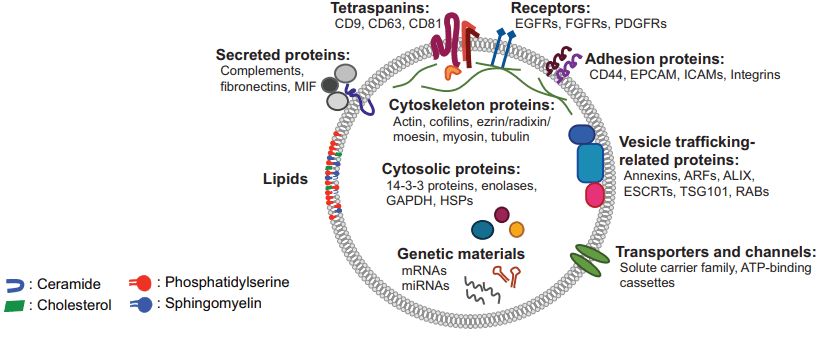 Fig 1. The rich content of mammalian extracellular vesicles. (Choi D S, et al. 2015)
Fig 1. The rich content of mammalian extracellular vesicles. (Choi D S, et al. 2015)
Exosomal Proteomics and LC-MS/MS Services
A detailed understanding of the biochemical (protein and lipid) composition of EV subgroups and the extent to which EV composition reflects the composition of source cells is necessary for the further development of diagnosis and treatment. We use liquid chromatography-tandem mass spectrometry (LC-MS/MS) to examine the exosomal proteome in the sample, and then select representative proteins with differences for gene ontology (GO) enrichment analysis. Protein extraction followed the same protocol as Western blotting, applying a certain amount of total protein to SDS-PAGE. Customers can also choose to send the gel fragments directly to our laboratory for mass spectrometry and data analysis. The data generated by mass spectrometry can be compared with the ExoCarta database. The surface protein content of exosomes can be used as a rich source of biomarkers and can provide potential surface marker targets for downstream antibody bead purification and FAC experiments. Exosomal isolates usually contain membrane proteins, especially four transmembrane proteins, and different amounts of extracellular matrix proteins. Microvesicle isolates may contain mitochondrial or endoplasmic reticulum-derived proteins. Our customized mass spectrometry service enables researchers to discover protein biomarkers on the surface and/or inside of exosomes.
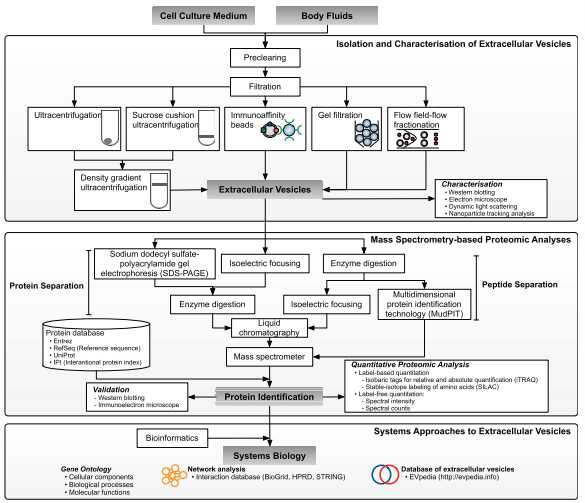 Fig 2. Overview of methodological approaches in high-throughput mass spectrometry-based proteomic analyses of extracellular vesicles. (Choi D S, et al. 2015)
Fig 2. Overview of methodological approaches in high-throughput mass spectrometry-based proteomic analyses of extracellular vesicles. (Choi D S, et al. 2015)
Exosomal Lipidomics and MS/MS Services
Similar to proteomics analysis, our lipidomics analysis service is also based on mass spectrometry. The exosomal lipid purification is carried out by the classic lipid extraction method (extraction with chloroform/methanol solution), and the sample is dried for MS/MS analysis. The detected lipid species are defined by the head group, number, length, and saturation of the fatty acid tail, and then further analysis is performed. The data mining of lipid profiles is narrower than that of proteomics. You can refer to the report of the ExoCarta database to screen the target of interest. Similarly, in-depth data mining includes cluster analysis and analysis of metabolic pathways.
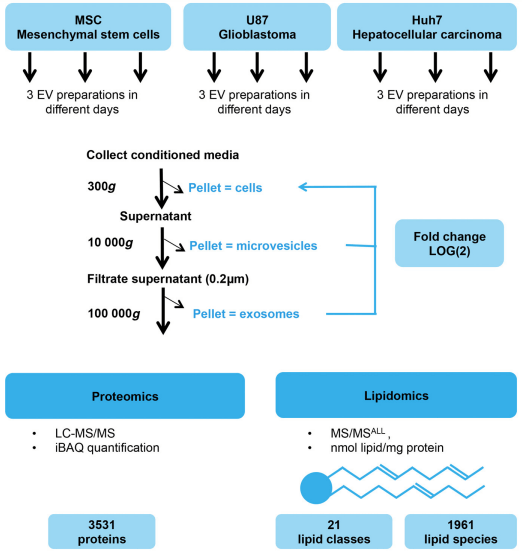 Fig 3. Workflow of EV preparation and mass spectrometry. (Haraszti R A, et al. 2016)
Fig 3. Workflow of EV preparation and mass spectrometry. (Haraszti R A, et al. 2016)
Creative Bioarray uses biochemical analysis and mass spectrometry methods to study the protein and lipid composition of EVs from various sources. These works provide the basis for the identification of biomarkers in EVs for research quality control purposes. With many years of exosome service experience, we are confident to provide customers with first-class mass spectrometry and data analysis services.If you are interested in our services or have any specific needs, please feel free to contact us. We look forward to working with you in the near future.
References:
- Choi D S, Kim D K, Kim Y K, et al. Proteomics of extracellular vesicles: exosomes and ectosomes[J]. Mass spectrometry reviews, 2015, 34(4): 474-490.
- Haraszti R A, Didiot M C, Sapp E, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources[J]. Journal of extracellular vesicles, 2016, 5(1): 32570.
For research use only. Not for any other purpose.

 Fig 1. The rich content of mammalian extracellular vesicles. (Choi D S, et al. 2015)
Fig 1. The rich content of mammalian extracellular vesicles. (Choi D S, et al. 2015)  Fig 2. Overview of methodological approaches in high-throughput mass spectrometry-based proteomic analyses of extracellular vesicles. (Choi D S, et al. 2015)
Fig 2. Overview of methodological approaches in high-throughput mass spectrometry-based proteomic analyses of extracellular vesicles. (Choi D S, et al. 2015)  Fig 3. Workflow of EV preparation and mass spectrometry. (Haraszti R A, et al. 2016)
Fig 3. Workflow of EV preparation and mass spectrometry. (Haraszti R A, et al. 2016) 